Precision Medicine: Predicting Anti-Cancer Drug Response
[Summary & Contributions] | [Relevant Publications]
Summary and Contributions
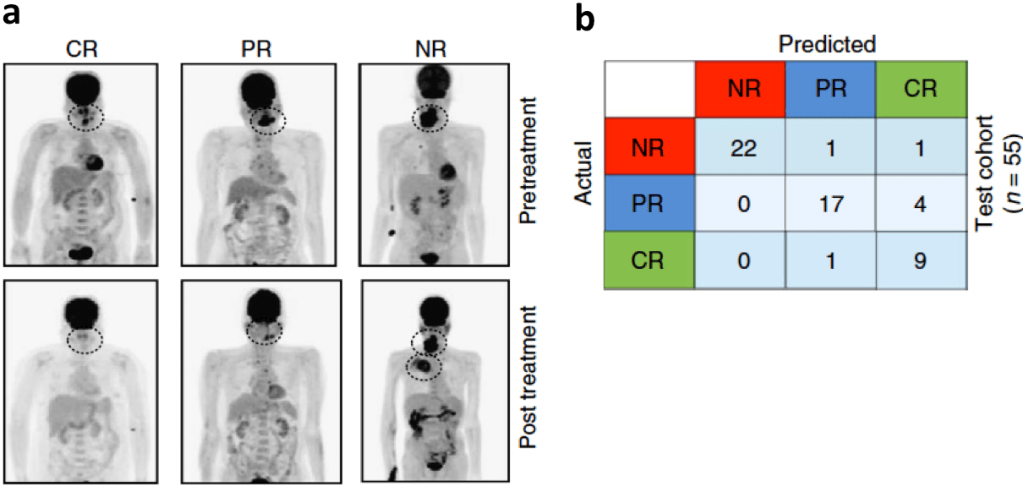
One of the problems that doctors treating patients with various complex diseases such as cancer often struggle with is in deciding which drug or treatment to administer to which patient: for many such diseases, patients with the same symptoms, genetic biomarkers, and other diagnostic test results, when given the same treatment, can present vastly different outcomes. Being able to better predict treatment outcomes is therefore critical to ensure that patients’ precious time and resources are spent on treatments to which they are most likely to respond well. We looked at this problem in collaboration with medical scientists; the goal of the collaboration was to design a model to predict whether a patient would show complete response (CR), partial response (PR), or no response (NR) to a given cancer treatment, based on training data associated with patients with head and neck squamous cell carcinoma (HNSCC) and with colorectal cancer. For each patient, we had a set of biological readouts from an ex vivo culture in which a biopsy sample from the patient’s tumor had been administered the drug under consideration, together with the clinical CR/PR/NR outcome observed when the same drug was actually given to the patient; these outcomes were used as labels in the training data, and for evaluating the learned model on the test data (see figure below). Since predicting a potential responder as a non-responder is more costly than prediction errors in the other direction, we used a variant of the SVMpAUC algorithm developed in our group to first learn a model that optimized the partial AUC up to a certain acceptable false positive rate treating CR, PR as positives and NR as negatives, and then refined the model to make 3-way predictions. For colorectal cancer, previous biomarker-based patient selection led to a positive response rate of ∼30%; for HNSCC, the previous positive response rate was ∼67%. Using our machine-learned patient selection approach led to a combined positive response rate of ∼94%.
Relevant Publications
- Biswanath Majumder, Ulaganathan Baraneedharan, Saravanan Thiyagarajan, Padhma Radhakrishnan, Harikrishna Narasimhan, Muthu Dhandapani, Nilesh Brijwani, Dency D. Pinto, Arun Prasath, Basavaraja U. Shanthappa, Allen Thayakumar, Rajagopalan Surendran, Govind K. Babu, Ashok M. Shenoy, Moni A. Kuriakose, Guillaume Bergthold, Peleg Horowitz, Massimo Loda, Rameen Beroukhim, Shivani Agarwal, Shiladitya Sengupta, Mallikarjun Sundaram and Pradip K. Majumder.
Predicting clinical response to anticancer drugs using an exvivo platform that captures tumor heterogeneity.
Nature Communications, 6:6169, 2015.
[pdf]